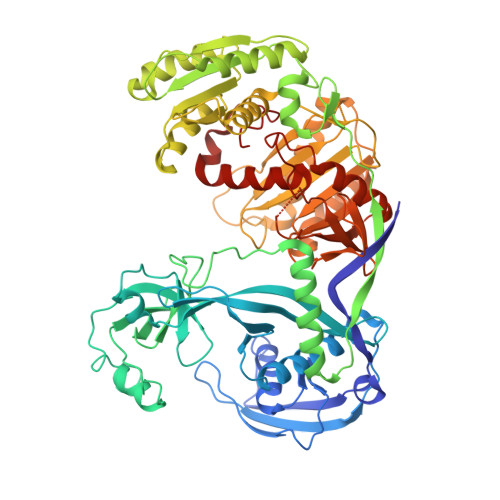
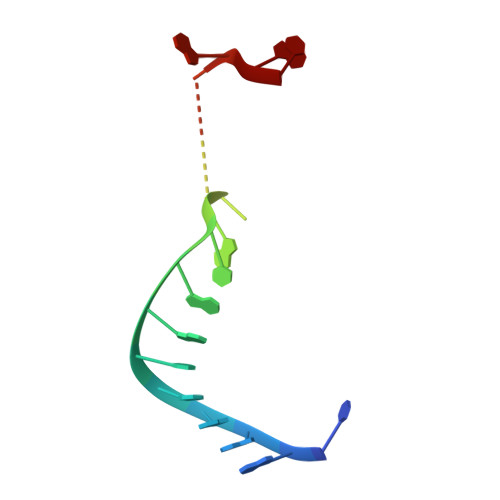
Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.
Wang, Y., Juranek, S., Li, H., Sheng, G., Tuschl, T., Patel, D.J.(2008) Nature 456: 921-926
- PubMed: 19092929
- DOI: https://doi.org/10.1038/nature07666
- Primary Citation of Related Structures:
3F73 - PubMed Abstract:
Here we report on a 3.0 A crystal structure of a ternary complex of wild-type Thermus thermophilus argonaute bound to a 5'-phosphorylated 21-nucleotide guide DNA and a 20-nucleotide target RNA containing cleavage-preventing mismatches at the 10-11 step. The seed segment (positions 2 to 8) adopts an A-helical-like Watson-Crick paired duplex, with both ends of the guide strand anchored in the complex. An arginine, inserted between guide-strand bases 10 and 11 in the binary complex, locking it in an inactive conformation, is released on ternary complex formation. The nucleic-acid-binding channel between the PAZ- and PIWI-containing lobes of argonaute widens on formation of a more open ternary complex. The relationship of structure to function was established by determining cleavage activity of ternary complexes containing position-dependent base mismatch, bulge and 2'-O-methyl modifications. Consistent with the geometry of the ternary complex, bulges residing in the seed segments of the target, but not the guide strand, were better accommodated and their complexes were catalytically active.
Organizational Affiliation:
Structural Biology Program, Memorial-Sloan Kettering Cancer Center, New York, New York 10065, USA.