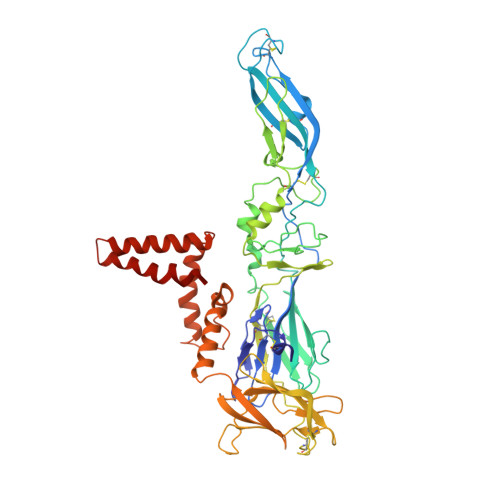
Structure of tick-borne encephalitis virus and its neutralization by a monoclonal antibody.
Fuzik, T., Formanova, P., Ruzek, D., Yoshii, K., Niedrig, M., Plevka, P.(2018) Nat Commun 9: 436-436
- PubMed: 29382836
- DOI: https://doi.org/10.1038/s41467-018-02882-0
- Primary Citation of Related Structures:
5O6A, 5O6V - PubMed Abstract:
Tick-borne encephalitis virus (TBEV) causes 13,000 cases of human meningitis and encephalitis annually. However, the structure of the TBEV virion and its interactions with antibodies are unknown. Here, we present cryo-EM structures of the native TBEV virion and its complex with Fab fragments of neutralizing antibody 19/1786. Flavivirus genome delivery depends on membrane fusion that is triggered at low pH. The virion structure indicates that the repulsive interactions of histidine side chains, which become protonated at low pH, may contribute to the disruption of heterotetramers of the TBEV envelope and membrane proteins and induce detachment of the envelope protein ectodomains from the virus membrane. The Fab fragments bind to 120 out of the 180 envelope glycoproteins of the TBEV virion. Unlike most of the previously studied flavivirus-neutralizing antibodies, the Fab fragments do not lock the E-proteins in the native-like arrangement, but interfere with the process of virus-induced membrane fusion.
- Structural Virology, Central European Institute of Technology, Masaryk University, Kamenice 753/5, 62500, Brno, Czech Republic.
Organizational Affiliation: