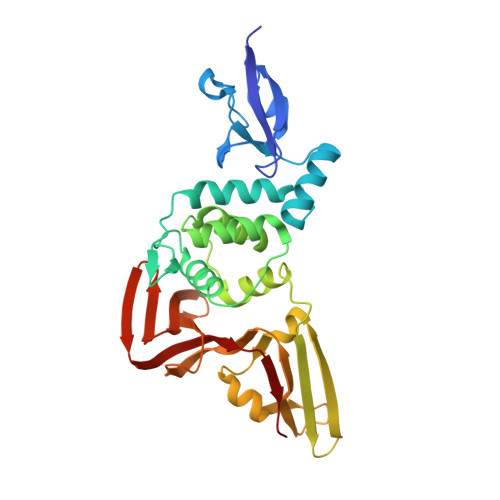
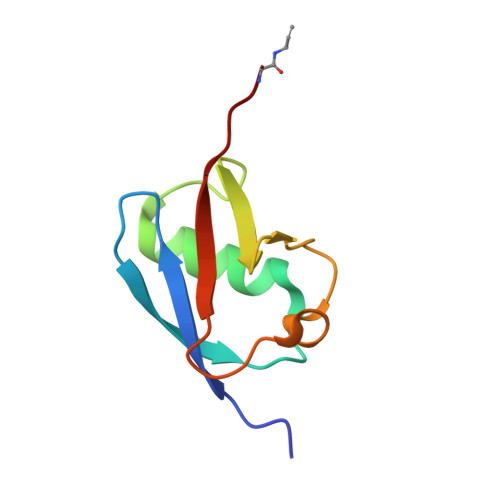
Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Klemm, T., Ebert, G., Calleja, D.J., Allison, C.C., Richardson, L.W., Bernardini, J.P., Lu, B.G., Kuchel, N.W., Grohmann, C., Shibata, Y., Gan, Z.Y., Cooney, J.P., Doerflinger, M., Au, A.E., Blackmore, T.R., van der Heden van Noort, G.J., Geurink, P.P., Ovaa, H., Newman, J., Riboldi-Tunnicliffe, A., Czabotar, P.E., Mitchell, J.P., Feltham, R., Lechtenberg, B.C., Lowes, K.N., Dewson, G., Pellegrini, M., Lessene, G., Komander, D.(2020) EMBO J 39: e106275-e106275
- PubMed: 32845033
- DOI: https://doi.org/10.15252/embj.2020106275
- Primary Citation of Related Structures:
6XA9, 6XAA - PubMed Abstract:
The SARS-CoV-2 coronavirus encodes an essential papain-like protease domain as part of its non-structural protein (nsp)-3, namely SARS2 PLpro, that cleaves the viral polyprotein, but also removes ubiquitin-like ISG15 protein modifications as well as, with lower activity, Lys48-linked polyubiquitin. Structures of PLpro bound to ubiquitin and ISG15 reveal that the S1 ubiquitin-binding site is responsible for high ISG15 activity, while the S2 binding site provides Lys48 chain specificity and cleavage efficiency. To identify PLpro inhibitors in a repurposing approach, screening of 3,727 unique approved drugs and clinical compounds against SARS2 PLpro identified no compounds that inhibited PLpro consistently or that could be validated in counterscreens. More promisingly, non-covalent small molecule SARS PLpro inhibitors also target SARS2 PLpro, prevent self-processing of nsp3 in cells and display high potency and excellent antiviral activity in a SARS-CoV-2 infection model.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research and Department of Medical Biology, University of Melbourne, Melbourne, Vic., Australia.