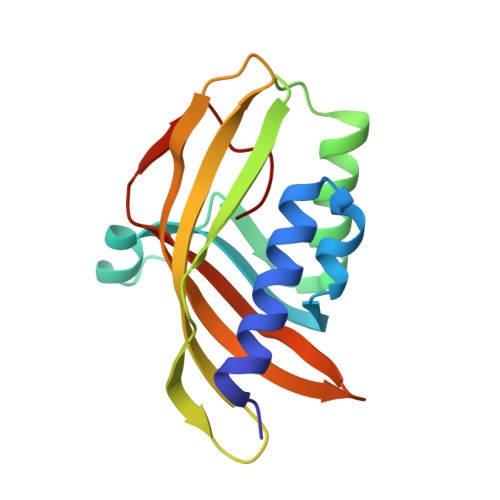
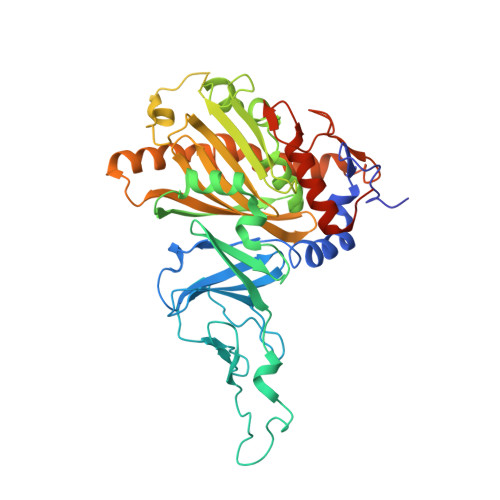
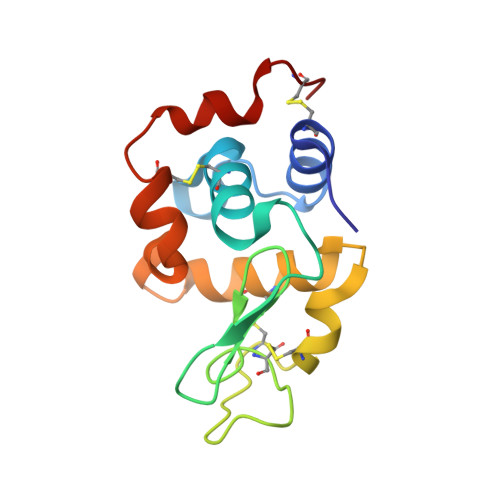
Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Kincannon, W.M., Zahn, M., Clare, R., Lusty Beech, J., Romberg, A., Larson, J., Bothner, B., Beckham, G.T., McGeehan, J.E., DuBois, J.L.(2022) Proc Natl Acad Sci U S A 119: e2121426119-e2121426119
- PubMed: 35312352
- DOI: https://doi.org/10.1073/pnas.2121426119
- Primary Citation of Related Structures:
7Q04, 7Q05, 7Q06 - PubMed Abstract:
SignificanceMore than 400 million tons of plastic waste is produced each year, the overwhelming majority of which ends up in landfills. Bioconversion strategies aimed at plastics have emerged as important components of enabling a circular economy for synthetic plastics, especially those that exhibit chemically similar linkages to those found in nature, such as polyesters. The enzyme system described in this work is essential for mineralization of the xenobiotic components of poly(ethylene terephthalate) (PET) in the biosphere. Our description of its structure and substrate preferences lays the groundwork for in vivo or ex vivo engineering of this system for PET upcycling.
Organizational Affiliation:
Department of Biochemistry, Montana State University, Bozeman, MT 59717.