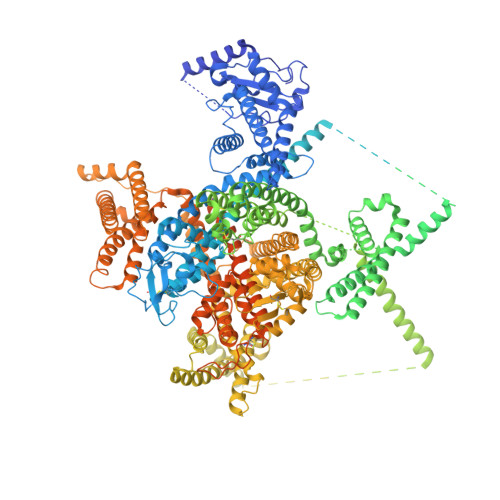
Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Huang, G., Wu, Q., Li, Z., Jin, X., Huang, X., Wu, T., Pan, X., Yan, N.(2022) Proc Natl Acad Sci U S A 119: e2209164119-e2209164119
- PubMed: 35878056
- DOI: https://doi.org/10.1073/pnas.2209164119
- Primary Citation of Related Structures:
7XVE, 7XVF - PubMed Abstract:
Voltage-gated sodium (Na v ) channel Na v 1.7 has been targeted for the development of nonaddictive pain killers. Structures of Na v 1.7 in distinct functional states will offer an advanced mechanistic understanding and aid drug discovery. Here we report the cryoelectron microscopy analysis of a human Na v 1.7 variant that, with 11 rationally introduced point mutations, has a markedly right-shifted activation voltage curve with V 1/2 reaching 69 mV. The voltage-sensing domain in the first repeat (VSD I ) in a 2.7-Å resolution structure displays a completely down (deactivated) conformation. Compared to the structure of WT Na v 1.7, three gating charge (GC) residues in VSD I are transferred to the cytosolic side through a combination of helix unwinding and spiral sliding of S4 I and ∼20° domain rotation. A conserved WNФФD motif on the cytoplasmic end of S3 I stabilizes the down conformation of VSD I . One GC residue is transferred in VSD II mainly through helix sliding. Accompanying GC transfer in VSD I and VSD II , rearrangement and contraction of the intracellular gate is achieved through concerted movements of adjacent segments, including S4-5 I , S4-5 II , S5 II , and all S6 segments. Our studies provide important insight into the electromechanical coupling mechanism of the single-chain voltage-gated ion channels and afford molecular interpretations for a number of pain-associated mutations whose pathogenic mechanism cannot be revealed from previously reported Na v structures.
Organizational Affiliation:
Westlake Laboratory of Life Sciences and Biomedicine, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou 310024, China.