Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism
Schneider, S., Kuehlbrandt, W., Yildiz, O.(2024) Structure
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Low-affinity phosphate transporter PHO90 | 881 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: PHO90, YJL198W, J0336 | 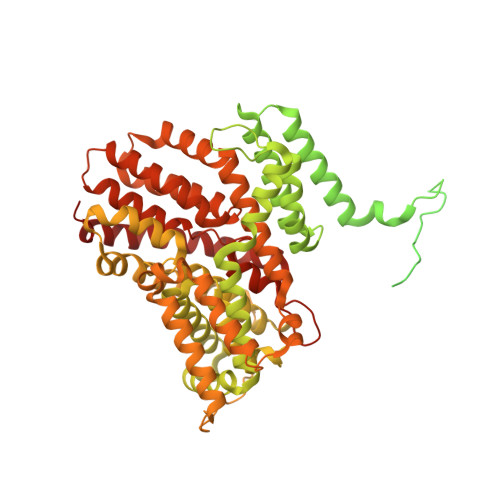 | |
UniProt | |||||
Find proteins for P39535 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P39535 Go to UniProtKB: P39535 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P39535 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 3PH Query on 3PH | F [auth A] G [auth A] H [auth A] I [auth A] J [auth B] | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE C39 H77 O8 P YFWHNAWEOZTIPI-DIPNUNPCSA-N |  | ||
| PO4 (Subject of Investigation/LOI) Query on PO4 | C [auth A], K [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| NA (Subject of Investigation/LOI) Query on NA | D [auth A], E [auth A], L [auth B], M [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | |
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Max Planck Society | Germany | -- |