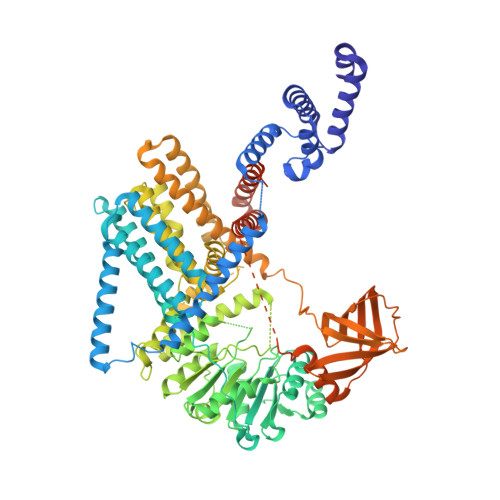
Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Anso, I., Zouhir, S., Sana, T.G., Krasteva, P.V.(2024) Nat Commun 15: 8799-8799
- PubMed: 39394223
- DOI: https://doi.org/10.1038/s41467-024-53113-8
- Primary Citation of Related Structures:
9FMT, 9FMV, 9FMZ, 9FNN, 9FO7, 9FP0, 9FP2 - PubMed Abstract:
Bacterial cellulosic polymers constitute a prevalent class of biofilm matrix exopolysaccharides that are synthesized by several types of bacterial cellulose secretion (Bcs) systems, which include conserved cyclic diguanylate (c-di-GMP)-dependent cellulose synthase modules together with diverse accessory subunits. In E. coli, the biogenesis of phosphoethanolamine (pEtN)-modified cellulose relies on the BcsRQABEFG macrocomplex, encompassing inner-membrane and cytosolic subunits, and an outer membrane porin, BcsC. Here, we use cryogenic electron microscopy to shed light on the molecular mechanisms of BcsA-dependent recruitment and stabilization of a trimeric BcsG pEtN-transferase for polymer modification, and a dimeric BcsF-dependent recruitment of an otherwise cytosolic BcsE 2 R 2 Q 2 regulatory complex. We further demonstrate that BcsE, a secondary c-di-GMP sensor, can remain dinucleotide-bound and retain the essential-for-secretion BcsRQ partners onto the synthase even in the absence of direct c-di-GMP-synthase complexation, likely lowering the threshold for c-di-GMP-dependent synthase activation. Such activation-by-proxy mechanism could allow Bcs secretion system activity even in the absence of substantial intracellular c-di-GMP increase, and is reminiscent of other widespread synthase-dependent polysaccharide secretion systems where dinucleotide sensing and/or synthase stabilization are carried out by key co-polymerase subunits.
Organizational Affiliation:
Univ. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, F-33600, Pessac, France.