News
Access New Website Features
04/28
New and enhanced features have been added to the RCSB PDB resource:
- Download data in batches. The Download Tool can download groups of structures, experimental data files, sequences, and ligands in a variety of formats. This tool is launched as a stand-alone application using the Java Web Start protocol.
- Map genomic position to protein. Mutations in a gene can have profound effects on the function of a protein. This analysis tool highlights the location of a gene location (i.e., the site of a SNP).
- Explore sequence, structure, and related annotations. Protein Feature View provides a graphical summary of how sequence data corresponds to PDB entries. New options include visualization of sequence motifs and access to related organisms with the same gene name.
- Access large structures. "Large structures" (containing > 62 chains and/or 99999 ATOM lines) are represented as single files in PDBx/mmCIF and PDBML formats. These entries can be easily found (or omitted) using Advanced Search. Corresponding tar files containing minimal/best effort files in PDB format that represent these entries can be accessed through individual Structure Summary pages and using the Download Tool.
- Visualize structures quickly. JSmol (JavaScript Jmol), is now the default 3D viewer to improve usage speed. The new program PV (Protein Viewer) is also offered as a powerful visualization tool. For large structures that have traditionally rendered slowly or were unable to load in 3D viewers, new features have been implemented to improve visualization.
See the What's New page for more new features and examples.
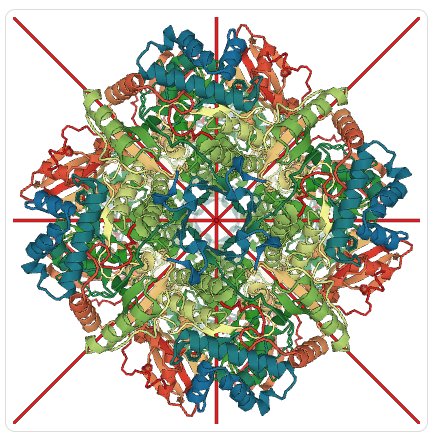
View of rubisco entry 4F0H oriented along symmetry axes (red tubes) using PV.












