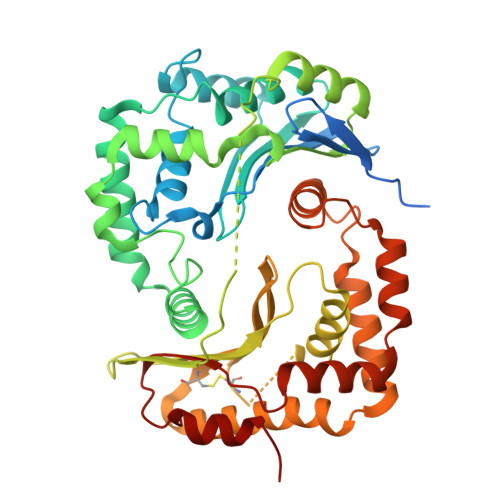
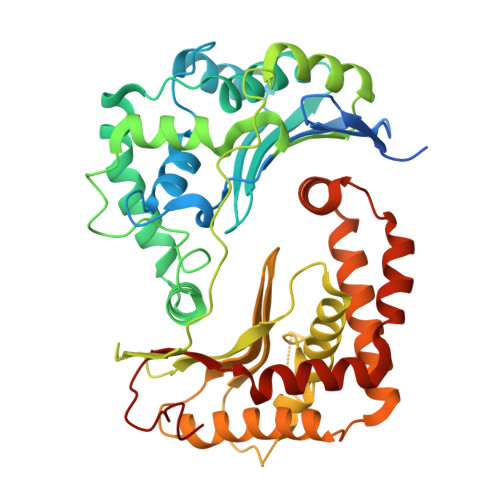
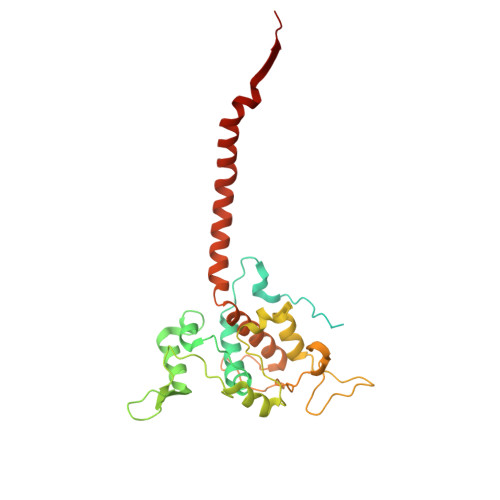
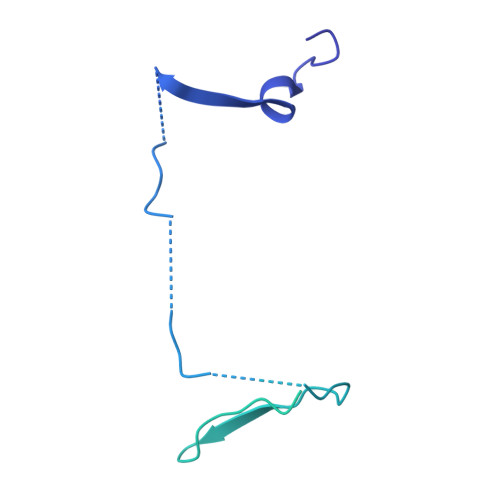
Structural basis of respiratory complex adaptation to cold temperatures.
Shin, Y.C., Latorre-Muro, P., Djurabekova, A., Zdorevskyi, O., Bennett, C.F., Burger, N., Song, K., Xu, C., Paulo, J.A., Gygi, S.P., Sharma, V., Liao, M., Puigserver, P.(2024) Cell 187: 6584
- PubMed: 39395414
- DOI: https://doi.org/10.1016/j.cell.2024.09.029
- Primary Citation of Related Structures:
8IAO, 8IAP, 8IAQ, 8IAR, 8IB4, 8IB5, 8IB6, 8IB7, 8IB9, 8IBA, 8IBB, 8IBC, 8IBD, 8IBE, 8IBF, 8IBG, 8IC2, 8IC3, 8IC4, 8IC5, 8XNL, 8XNM, 8XNN, 8XNO, 8XNP, 8XNQ, 8XNR, 8XNS, 8XNT, 8XNU, 8XNV, 8XNW, 8XNX, 8XNY, 8XNZ, 8XO0 - PubMed Abstract:
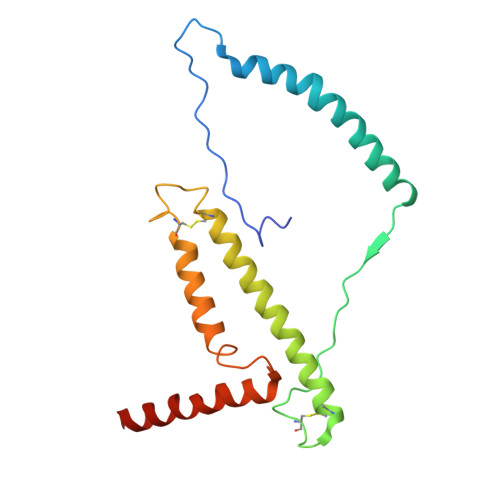
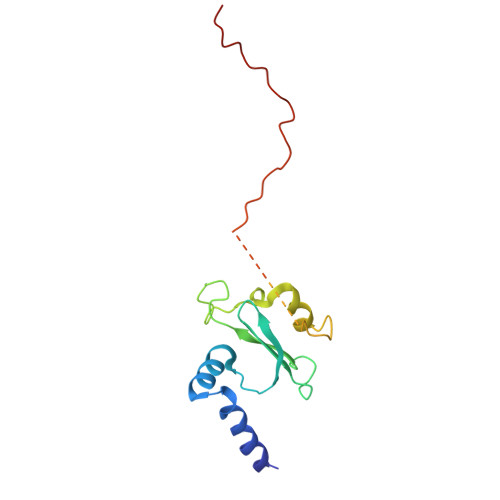
In response to cold, mammals activate brown fat for respiratory-dependent thermogenesis reliant on the electron transport chain. Yet, the structural basis of respiratory complex adaptation upon cold exposure remains elusive. Herein, we combined thermoregulatory physiology and cryoelectron microscopy (cryo-EM) to study endogenous respiratory supercomplexes from mice exposed to different temperatures. A cold-induced conformation of CI:III 2 (termed type 2) supercomplex was identified with a ∼25° rotation of CIII 2 around its inter-dimer axis, shortening inter-complex Q exchange space, and exhibiting catalytic states that favor electron transfer. Large-scale supercomplex simulations in mitochondrial membranes reveal how lipid-protein arrangements stabilize type 2 complexes to enhance catalytic activity. Together, our cryo-EM studies, multiscale simulations, and biochemical analyses unveil the thermoregulatory mechanisms and dynamics of increased respiratory capacity in brown fat at the structural and energetic level.
Organizational Affiliation:
Department of Chemical Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, China; Department of Cell Biology, Harvard Medical School, Boston, MA 02115, USA.